D-cycloserine was restructured to treat tuberculosis
D-cycloserine was restructured to treat tuberculosis
December 08, 2017 Source: Sina Pharmaceutical
Window._bd_share_config={ "common":{ "bdSnsKey":{ },"bdText":"","bdMini":"2","bdMiniList":false,"bdPic":"","bdStyle":" 0","bdSize":"16"},"share":{ }};with(document)0[(getElementsByTagName('head')[0]||body).appendChild(createElement('script')) .src='http://bdimg.share.baidu.com/static/api/js/share.js?v=89860593.js?cdnversion='+~(-new Date()/36e5)];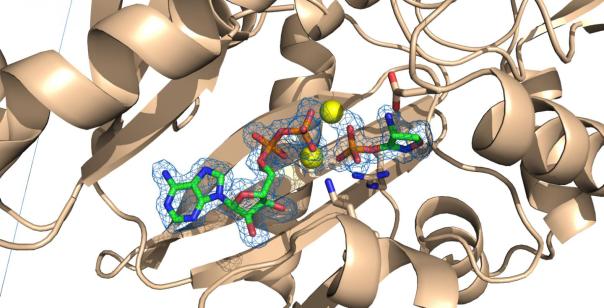
â–² Phosphorylated D-cycloserine binds to the complex site of magnesium ion and ADP to the active site of E. coli D-alanine-D alanine ligase
Tuberculosis (TB) remains a global public health threat that is also the leading cause of death worldwide. Therefore, it is essential to find new drugs that effectively control and treat tuberculosis.
A new study by scientists at the University of Warwick and the Francis Crick Institute in the UK will help to more effectively address tuberculosis and other life-threatening microbial infections. The disease, and all of this, is attributed to an ancient antibiotic: D-cylcoserine. The study was recently published online in the top journal Nature Communication, titled: Inhibition of D-Ala: D-Ala Ligase through a Phosphorylated Form of the Antibiotic D-Cycloserine (phosphorylated form) D-cycloserine inhibits D alanine: D alanine ligase), and the findings provide important insights into the design of new antibiotics.
D-cycloserine is an ancient antibiotic that can effectively treat a variety of diseases caused by microbial infections, including tuberculosis, but this drug is often used for second-line treatment due to some side effects.
Interestingly, the researchers found that the antibiotic can act on multiple bacterial targets in a variety of different chemical ways, which is probably the only antibiotic in the world that can act in this way. D-cycloserine attacks bacteria by inhibiting two different enzymes (D-alanine racemase and D-alanine-D alanine ligase), each of which establishes and maintains bacteria Required for structural integrity of the cell wall. In the case of D-alanine racemase, D-cycloserine is capable of forming a molecular bond with a chemical group required for enzymatic activity, which stops the enzyme.
Professor David Roper, the author of the study, said that in this new discovery, we observed that D-cycloserine binds to D-alanine-D alanine ligase and chemically modifies the enzyme to form a chemical species. I have never seen it before. We now fully understand how this antibiotic works in a completely different way for different targets, which may be unique among the antibiotic family. The researchers first observed how D-cycloserine inhibits D-alanine-D-alanine ligase.
Dr. Luiz Pedro Carvolho, head of the Mycobacterium tuberculosis metabolism and antibiotics research group at the Francis Crick Institute, said that perhaps more important than how D-cycloserine works, the study highlights an increasingly obvious fact: Our understanding of the true mode of action of antibiotics is far less than we think. Only by truly understanding the molecular or cellular events triggered by antibiotics or reacting to the presence of antibiotics can we truly understand how to improve drugs, which is urgently needed for the current threat of antibiotic resistance.
The team noted that its long-term goal is to modify the structure of D-cycloserine to bring it closer to newly discovered chemicals, thus developing a new antibiotic that is more specific and avoids the side effects of D-cycloserine. To make it more widely used in the treatment of antibiotic-resistant infections.
With the increasing severity of current antibiotic resistance, there is a global crisis in the field of infectious disease treatment. Antibiotic resistance threatens all aspects of human activity, including medicine and agriculture. Antibiotic resistance may lead to more deaths than cancer. (Sina Pharmaceutical Compilation/newborn)
Article reference source:
1, Old Antibiotic Repurposed to Treat TB
2. Inhibition of D-Ala: D-Ala ligase through a phosphorylated form of the antibiotic D-cycloserine
Original title: Ancient antibiotics rejuvenated youth D-cycloserine was restructured to treat tuberculosis
Sanger Sequencing, which was used for DNA nucleotide sequencing, was created and rewarded Nobel Prize by Frederick Sanger in 1977.
How does the Sanger Sequencing work?
Firstly, we use the primer to have a short primer binding next to the region of interest on the DNA template. Then, the DNA polymerase starts building up from the primer by adding complementary nucleotides to the DNA strand.
To find the STOP that allows us to identify the base of the very end of the particular DNA fragment. Sanger sequencing principle is applied to removing an oxygen atom from the ribonucleotide. ddNTP (Dideoxynucleotide) is to put a wrench into a gear. The polymerase enzyme will no longer add normal nucleotides to this DNA chain. The extension would stop.
We can identify the Chain terminating nucleotide by a specific fluorescent dye. Superyears Genetic Analyzer can support fluorescent dye up to 6/8 colors to be exact.
Sanger sequencing results in the formation of extension products of various lengths terminated with dideoxynucleotides at the 3' end.
Capillary Electrophoresis separates the extension products.
An electrical current injects the molecules into a long capillary tube filled with a gel polymer.
During CE (Capillary Electrophoresis), negatively charged DNA fragments move forward to the Positive electrode.
The speed at which a DNA fragment migrates through the medium is inversely proportional to its molecular weight.
This process can separate the extension products by size at a resolution of one base.
A laser excites the dye-labeled DNA fragments as they pass through a tiny window at the end of the capillary.
The excited dye emits light at a characteristic wavelength that is detected by a light sensor.
The software can interpret the detected signal and translate it into a basecall.
A sequencing reaction is performed in the presence of all four terminated nucleotides, a pool of DNA fragments that are measured and separated base by base.
Data file showing the sequence of the DNA in a colorful electropherogram.
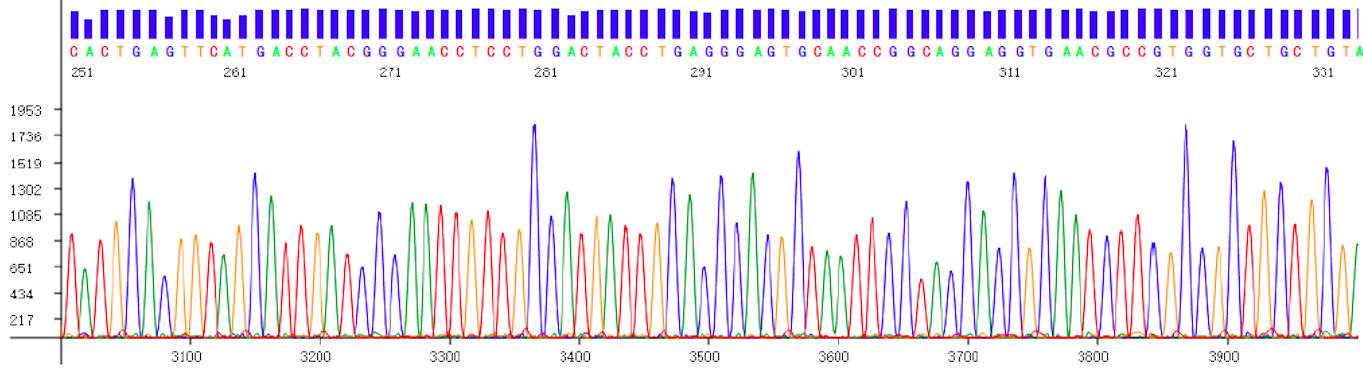
Based on the Sanger sequencing principle, the gene analyzer produced by Superyears Gene can be used for gene sequence analysis and fragment analysis, which can quickly detect multi-target (10-70 loci) genes at a time. The Genetic Analyzer has the advantages of low single detection cost, simple operation, and intuitive and straightforward result interpretation. It is a powerful tool for medium-throughput and multi-gene joint inspection.
Superyears focus on developing a genetic instrument that brings the best testing results. We can partner with you to create better solutions with our ongoing efforts and innovation that support laboratory productivity. Superyears gene company can provide high-quality and value-added products that can generate excellent outcomes from lab testing. Including DNA extractor for samples collection, Real-time PCR for samples amplification, compatible reagents within the use of multiple mainstream instruments, state-of-art genetic testing analyzer, and self-sufficient analyzing software.
Superyears Gene Technology company provides Classic series genetic analyzer based on Sanger Sequencing Principle. Superyears has made a breakthrough with the state-of-the-art Eight-color Fluorescence technology and optional 8-,16-,24-,96- (under research) channel analyzer, which can be applied in gene sequencing and fragment analysis. Therefore, it is suitable for fundamental Molecular Genetic Research, Clinical Medicine, Food Safety, Agricultural Science, and other scenarios. In addition, classic 116 has been approved by NMPA and can be used for in vitro diagnosis."
Sanger Sequecing Instrument,Genetic Analyzer Forensic Dna Instrument,Sanger Sequecing Dna Analyzer,Sanger Sequencing Method
Nanjing Superyears Gene Technology Co., Ltd. , https://www.superyearsglobal.com